Abstract
Background: SARS-Cov-2 infections are associated with increased mortality and morbidity, largely due to inflammatory cascades and cytokine release syndrome (CRS). Clonal hematopoiesis of indeterminate potential (CHIP) is defined by the presence and subsequent expansion of somatic, leukemia-associated driver mutations in apparently healthy individuals with normal blood counts. CHIP has been associated with increased inflammation, with cytokines such as IL1-b, IL6 and TNF-a being elevated at baseline in affected individuals. We hypothesized that the presence of CHIP in patients with COVID-19 would result in excessive inflammation-related mortality and morbidity.
Methods: We used the Mayo Clinic COVID-19 database to identify patients with COVID-19 on whom peripheral blood mononuclear cells (PBMC) were available for research use (IRB approved). We carried out target-capture next generation sequencing for 220 CH related genes, by previously described methods (1000 x coverage, variant allele fraction/VAF detection limit >0.5%; Kusne Y et al AJH 2021). CHIP was defined by the presence of a CH mutation with a VAF>1% in an individual with normal baseline blood counts. Demographics, blood counts, and inflammatory markers (CRP and cytokine levels- ELISA assay) at COVID-19 diagnosis and during follow-up (as clinically indicated) were collected. COVID-19 disease severity was classified based on the presence and severity of CRS, graded using the Penn Grading Scale (Porter et. al., 2018), and the WHO ordinal scale (WHO Blueprint, 2020). We used Fisher's exact test and the Wilcoxon rank sum test to compare categorical and continuous variables. Survival analysis was performed using the Kaplan-Meier method. We accounted for differences in age and sex using multivariable-adjusted proportional hazards regression models.
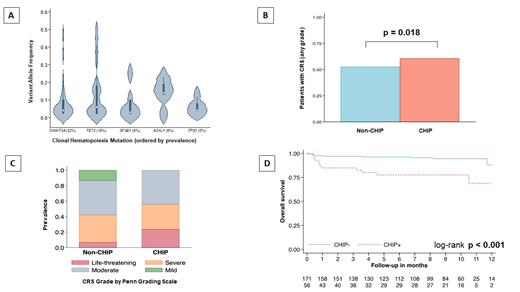
Results: Seventy-two CHIP mutations were detected in 56 (25%) of the 227 patients with COVID-19 that had PBMC available; median age 69 years (range; 42-99 years), 61% male. Fifteen (26%) patients had 2 CHIP mutations, while 1 patient had 3 CHIP mutations. Common mutations encountered included DNMT3A (32%) , TET2 (19%) , SF3B1 (8%) , ASXL1 (6%) , MPL (5%) , and TP53 (5%; Figure 1A). COVID-19 patients with CHIP were older in age (median 69 vs 57 years; p<0.0001) and had higher baseline MCP-1 (p=0.04) levels. However, there were no differences in sex, comorbidities, blood counts, IL1-b, IL6 and TNF-a levels between the two groups. The median follow-up for the entire cohort was 9 months. The relative change from baseline in blood counts and inflammatory markers (CRP and cytokines) during follow-up was similar in CHIP and non-CHIP patients, with the exception that COVID-19-onset neutropenia was more common in CHIP patients (8% vs 1%; p=0.017) compared to those without CHIP. At last follow up neutropenia had resolved in all patients.
Both groups had comparable number of patients with CRS (61% CHIP vs 53% non-CHIP patients, p=0.354, Figure 1B), however, CHIP patients had more severe CRS (median Penn Grade 3 versus 2 in non-CHIP, p=0.018, Figure 1C). Based on the WHO ordinal scale, CHIP patients were more likely to experience hospitalization with severe disease and death (61% versus 45% in non-CHIP, p = 0.049). Moreover, COVID-19 CHIP patients experienced worse overall survival in comparison to patients without CHIP (median 13.1 months vs not reached, p<0.001, Figure 1D). This association remained consistent after adjusting for age and sex at the time of COVID-19 diagnosis (HR 3.15, 95% 1.32 - 7.55, p = 0.010). At last follow-up, 22 deaths were documented: 13 (23%) in patients with CHIP and 9 (5%) in the non-CHIP group (p=0.02), with the primary cause for mortality being hypoxic respiratory failure (62% in CHIP vs 44% non-CHIP, p=0.04).
Conclusions: In this study, we observe an age-independent impact of CHIP on COVID-19 associated inflammatory morbidity (CRS) and mortality (hypoxemic respiratory failure). We are currently carrying out detailed single cell (ssDNA, RNA and ATAC-seq) and proteomic studies (O-link PEA assays) to better elucidate this pathophysiology.
Patnaik: Kura Oncology: Research Funding; StemLine: Research Funding.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal